Proton Recoil Telescope analysis#
Download the data#
First let’s download the data from the BABY-1L-run-3 available on Zenodo: https://zenodo.org/records/15177190
[1]:
import requests
from pathlib import Path
# Path to save the extracted files
output_file = Path("raw_2025-03-18_ROSY.h5")
if output_file.exists():
print(f"File already exists: {output_file}")
else:
# URL of the file
url = "https://zenodo.org/records/15177190/files/raw_2025-03-18%20ROSY.h5?download=1"
# Download the file
response = requests.get(url)
if response.status_code == 200:
print("Download successful!")
# Save the file to the specified directory
with open(output_file, "wb") as f:
f.write(response.content)
print(f"File saved to: {output_file}")
else:
print(f"Failed to download file. HTTP Status Code: {response.status_code}")
File already exists: raw_2025-03-18_ROSY.h5
Load data#
To load the data, we use the load_data_from_file function in the prt module of libra-toolbox.
[2]:
from libra_toolbox.neutron_detection.diamond import prt
data = prt.load_data_from_file(output_file)
['Channel A', 'Channel B', 'Channel C', 'Channel D', 'Coincidence']
Active channels: [ True True True True]
Channel 0: Channel A
Channel 1: Channel B
Channel 2: Channel C
Channel 3: Channel D
data is a dictionary containing, for each active channel, the timestamps in seconds and the amplitudes of the events in mV
[3]:
data
[3]:
{'Channel A': {'timestamps': array([ 415.78811039, 1072.84300216, 1073.88120264, ...,
42680.1049604 , 42680.34160139, 42680.55178646], shape=(296633,)),
'amplitudes': array([ 27.77429243, 27.26467238, 433.94146794, ..., 1119.63523789,
31.34163274, 403.87388531], shape=(296633,))},
'Channel B': {'timestamps': array([ 44.26180857, 112.7965396 , 137.2218714 , ...,
42681.64475139, 42681.64734641, 42681.6553663 ], shape=(4625419,)),
'amplitudes': array([ 31.26355419, 52.86844935, 77.77762261, ..., 237.9080221 ,
33.04278085, 530.71789544], shape=(4625419,))},
'Channel C': {'timestamps': array([1.12628682e+01, 4.42618085e+01, 8.61230073e+01, ...,
4.26819807e+04, 4.26819854e+04, 4.26819877e+04], shape=(4742660,)),
'amplitudes': array([52.92923246, 41.53298624, 30.64323984, ..., 36.72123783,
70.65672658, 39.00048708], shape=(4742660,))},
'Channel D': {'timestamps': array([1.49495708e+01, 3.27648884e+01, 6.77186525e+01, ...,
4.26822708e+04, 4.26822903e+04, 4.26822947e+04], shape=(4496082,)),
'amplitudes': array([ 37.03524888, 32.1823542 , 61.04430677, ..., 76.36923734,
40.86648152, 322.334373 ], shape=(4496082,))}}
[4]:
for channel, channel_data in data.items():
print(f"Found {len(channel_data["timestamps"])} ({len(channel_data["timestamps"]):.2e}) events in channel {channel}")
Found 296633 (2.97e+05) events in channel Channel A
Found 4625419 (4.63e+06) events in channel Channel B
Found 4742660 (4.74e+06) events in channel Channel C
Found 4496082 (4.50e+06) events in channel Channel D
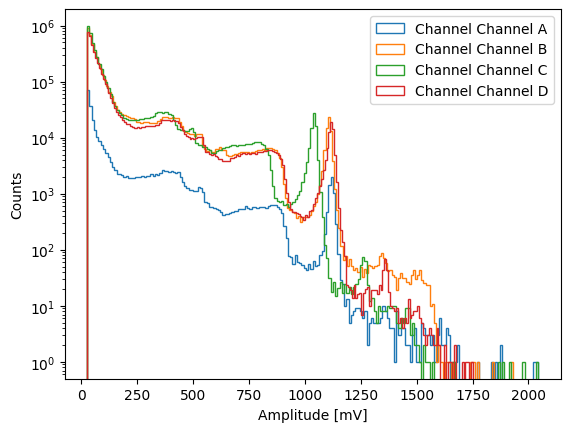
Channel A contains about 10 times less counts because diamond A is thinner than diamonds B, C, and D.
Plot data#
We can now plot the amplitudes of the events (analagous to energy, but uncalibrated)
[5]:
import matplotlib.pyplot as plt
for channel_name, channel_data in data.items():
plt.hist(
channel_data["amplitudes"],
bins=200,
histtype="step",
label=f"Channel {channel_name}",
)
plt.xlabel("Amplitude [mV]")
plt.ylabel("Counts")
plt.yscale("log")
plt.legend()
plt.show()
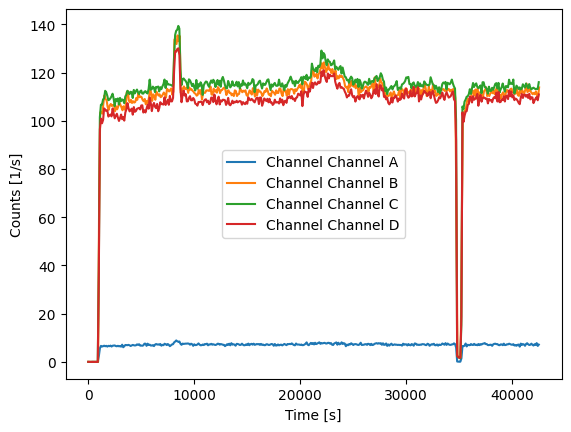
Calculate count rate#
With get_count_rate we can obtain the temporal evolution of the count rate (in count/s).
[6]:
for channel_name, channel_data in data.items():
count_rates, count_rate_bins = prt.get_count_rate(channel_data["timestamps"], bin_time=100)
plt.plot(count_rate_bins[:-1], count_rates, label=f"Channel {channel_name}")
plt.xlabel("Time [s]")
plt.ylabel("Counts [1/s]")
plt.legend()
plt.show()
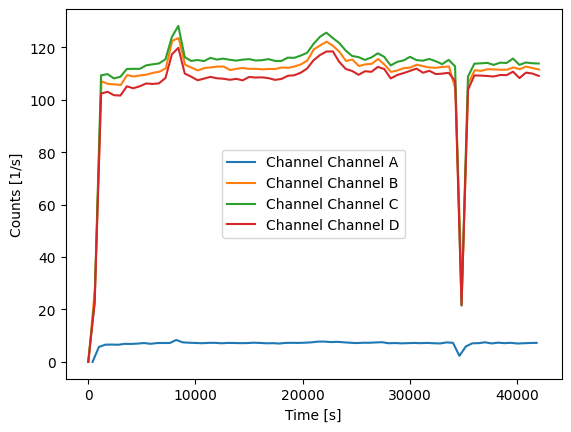
Increasing the bin_time will result in a smoother plot:
[7]:
for channel_name, channel_data in data.items():
count_rates, count_rate_bins = prt.get_count_rate(channel_data["timestamps"], bin_time=600)
plt.plot(count_rate_bins[:-1], count_rates, label=f"Channel {channel_name}")
plt.xlabel("Time [s]")
plt.ylabel("Counts [1/s]")
plt.legend()
plt.show()
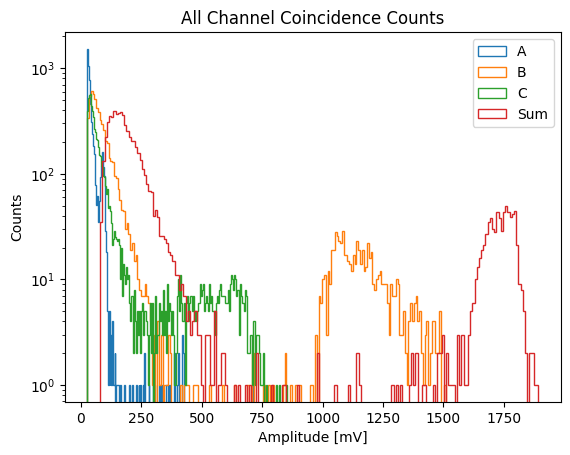
Plotting coincidences.#
The separate channels show energy deposited in an individual diamond by a given coincidence event, and the combined plot shows the total energy of a coincidence event found by summing the energy deposited in each diamond by the particle
[8]:
# Only the coincidence window (in seconds) and the coincidence_criteria needs to be changed. Everythign else is automatically done
# coincidence_window in seconds (1e-9 = 1 ns)
# coincidence_citeria:
# 0: Ignore thsi channel for the calculation
# 1: use this channel for coincidence caluclations
# 2: Use this channel for anti-coincidence (no count in the time window for this channel is allowed)
# -> structure [Channel A, Channel B, Channel C, Channel D]
df = prt.calculate_coincidence(
A_time=data["Channel A"]["timestamps"],
A_ampl=data["Channel A"]["amplitudes"],
B_time=data["Channel B"]["timestamps"],
B_ampl=data["Channel B"]["amplitudes"],
C_time=data["Channel C"]["timestamps"],
C_ampl=data["Channel C"]["amplitudes"],
D_time=data["Channel D"]["timestamps"],
D_ampl=data["Channel D"]["amplitudes"],
coincidence_window=100e-9, # 100 ns
coincidence_citeria=[1, 1, 1, 0],
)
Ignore: 1, Coincidence: 3, Anti-Coincidence: 0
[9]:
df.head()
[9]:
| A_time [s] | A_amplitude [mV] | B_time [s] | B_amplitude [mV] | C_time [s] | C_amplitude [mV] | Sum_amplitude [mV] | |
|---|---|---|---|---|---|---|---|
| 0 | 1073.983941 | 43.062894 | 1073.983941 | 39.142987 | 1073.983941 | 30.136740 | 112.342620 |
| 1 | 1099.970165 | 72.620856 | 1099.970165 | 1383.729992 | 1099.970165 | 66.098228 | 1522.449076 |
| 2 | 1102.823226 | 31.596443 | 1102.823226 | 53.376800 | 1102.823226 | 97.754468 | 182.727710 |
| 3 | 1106.815829 | 47.139854 | 1106.815829 | 156.571946 | 1106.815829 | 72.429476 | 276.141276 |
| 4 | 1107.332351 | 32.360873 | 1107.332351 | 45.751543 | 1107.332351 | 47.357734 | 125.470150 |
[10]:
print(f"Found {len(df)} coincidence events")
Found 6447 coincidence events
Channel D is not included because it was not used to calculate coincidences (14 MeV neutrons do not reach channel D)
[11]:
for label in ["A", "B", "C", "Sum"]:
plt.hist(
df[f"{label}_amplitude [mV]"],
bins=200,
histtype="step",
label=f"{label}",
)
plt.xlabel("Amplitude [mV]")
plt.ylabel("Counts")
plt.title("All Channel Coincidence Counts")
plt.yscale("log")
plt.legend()
plt.show()