Tritium release#
This example shows how to run a 0D tritium release model.
Theory#
The model is a simple ODE:
where \(V\) is the volume of the salt, \(c_\mathrm{salt}\) is the tritium concentration in the salt, \(S\) is the tritium source term, and \(Q_i\) are the different release rates.
The source term is expressed as:
where \(\mathrm{TBR}\) is the Tritium Breeding Ratio and \(\Gamma_n\) is the neutron rate.
The release rates are expressed as:
where \(A_i\) is the surface area, \(k_i\) is the mass transport coefficient, and \(c_\mathrm{external}\) is the external tritium concentration (assumed negligible compared to \(c_\mathrm{salt}\)).
Implementation#
First we import the right module:
[1]:
from libra_toolbox.tritium import model, ureg
We can then create a model using the Model class.
[2]:
import numpy as np
# geometry
baby_diameter = 1.77 * ureg.inches - 2 * 0.06 * ureg.inches # from CAD drawings
baby_volume = 0.100 * ureg.L
baby_radius = 0.5 * baby_diameter
baby_cross_section = np.pi * baby_radius**2
baby_height = baby_volume / baby_cross_section
# neutron rate
P383_neutron_rate = 4.95e8 / 2 * ureg.neutron * ureg.s**-1
A325_neutron_rate = 2.13e8 / 2 * ureg.neutron * ureg.s**-1
neutron_rate = P383_neutron_rate + A325_neutron_rate
# irradiation schedule
exposure_time = 12 * ureg.hour
irradiations = [
[0 * ureg.hour, 0 + exposure_time],
[24 * ureg.hour, 24 * ureg.hour + exposure_time],
]
my_model = model.Model(
radius=baby_radius,
height=baby_height,
TBR=5.4e-4 * ureg.particle * ureg.neutron**-1,
neutron_rate=neutron_rate,
k_top=1.6905e-6 * ureg.m * ureg.s**-1,
k_wall=5.0715e-8 * ureg.m * ureg.s**-1,
irradiations=irradiations,
)
Finally we run the model for 7 days:
[3]:
my_model.run(t_final=7 * ureg.days)
/home/remidm/miniconda3/envs/libra-toolbox/lib/python3.12/site-packages/scipy/integrate/_ivp/base.py:23: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
return np.asarray(fun(t, y), dtype=dtype)
Visualisation#
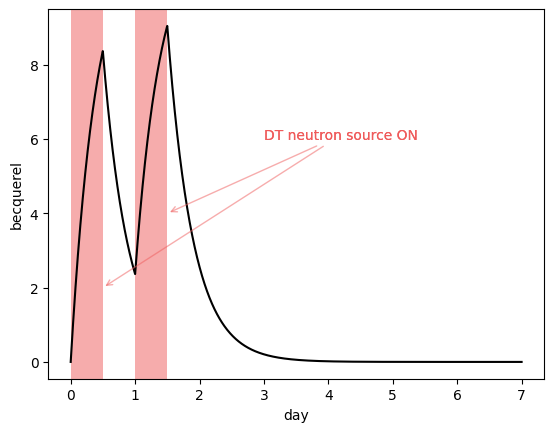
Salt tritium inventory#
The salt inventory \(I = V \ c_\mathrm{salt}\) can be plotted. The units are in Bq and computed from the specific activity of tritium.
[4]:
from libra_toolbox.tritium import plotting
import matplotlib.pyplot as plt
plotting.plot_salt_inventory(my_model, color="black")
plotting.plot_irradiation(my_model, facecolor="#EF5B5B", alpha=0.5)
plt.annotate(
"DT neutron source ON",
xy=(1.5 * ureg.day, 4 * ureg.Bq),
xytext=(3 * ureg.days, 6 * ureg.Bq),
color="#EF5B5B",
arrowprops=dict(arrowstyle="->", color="#EF5B5B", alpha=0.5),
)
plt.annotate(
"DT neutron source ON",
xy=(0.5 * ureg.day, 2 * ureg.Bq),
xytext=(3 * ureg.days, 6 * ureg.Bq),
color="#EF5B5B",
arrowprops=dict(arrowstyle="->", color="#EF5B5B", alpha=0.5),
)
plt.show()
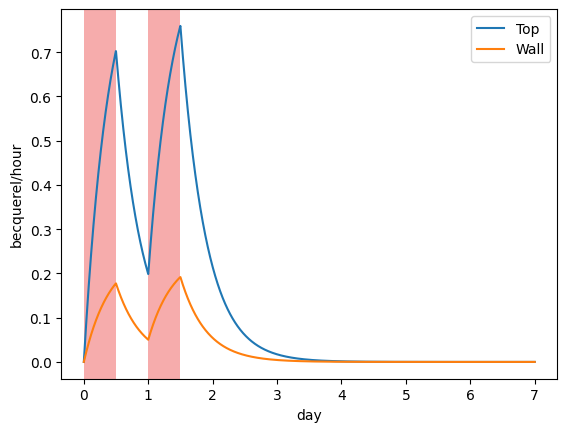
Release rates#
The release rates \(Q_i\) are computed and plotted:
[5]:
plotting.plot_top_release(my_model, label="Top")
plotting.plot_wall_release(my_model, label="Wall")
plotting.plot_irradiation(my_model, facecolor="#EF5B5B", alpha=0.5)
plt.legend()
plt.show()
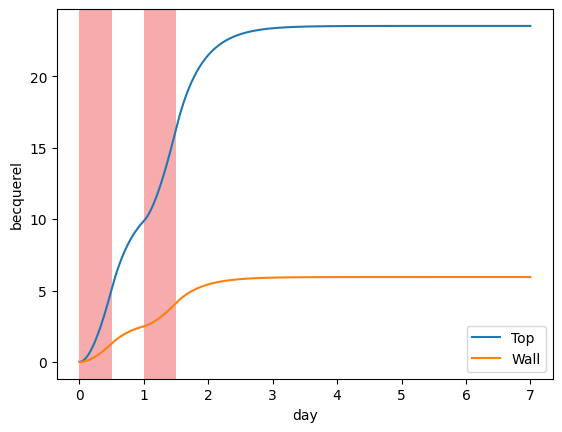
Cumulative releases#
It may be interesting to compute the cumulative release because this is what’s measured by the bubblers.
[6]:
plotting.plot_integrated_top_release(my_model, label="Top")
plotting.plot_integrated_wall_release(my_model, label="Wall")
plotting.plot_irradiation(my_model, facecolor="#EF5B5B", alpha=0.5)
plt.legend()
plt.show()
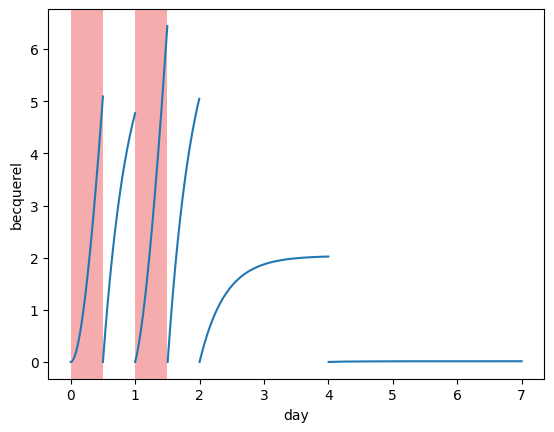
LSC sample activity#
When sampling, a sample of water is taken from the bubblers and its activity is measured in the Liquid Scintillation Counter.
The activity in the sample therefore depends on the collection volume (the bubbler vial) and the LSC sample volume (ie. how much water from the bubbler is being sampled).
If we assume that all of the water is replaced in the bubblers at each sampling time, then that means the water activity drops to zero.
Using plot_sample_activity_top we can try and predict what the sample activity is going to be given a list of sampling times.
[7]:
replacement_times = [0.5, 1, 1.5, 2, 4] * ureg.day
plotting.plot_sample_activity_top(my_model, replacement_times=replacement_times)
plotting.plot_irradiation(my_model, facecolor="#EF5B5B", alpha=0.5)
plt.show()
It is also possible to change the default collection volume and sample volume:
[8]:
for collection_vol in [10, 20, 100] * ureg.mL:
plotting.plot_sample_activity_top(
my_model,
replacement_times=replacement_times,
collection_vol=collection_vol,
label=f"{collection_vol:~.0f}",
)
plotting.plot_irradiation(my_model, facecolor="#EF5B5B", alpha=0.5)
plt.legend()
plt.show()
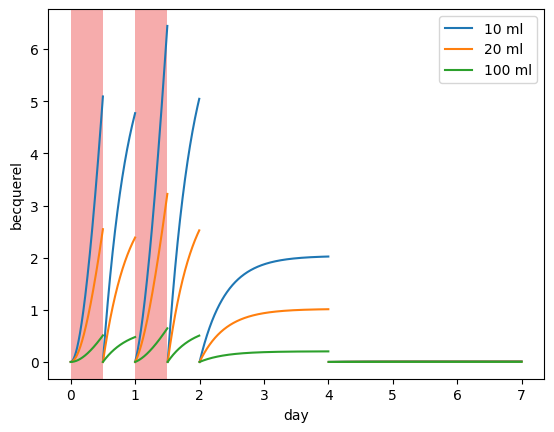
The larger the collection volume, the lower the sample activity (for a given sample volume). It is important to take this into account when designing an experiment and to compare with detectability limits.